Sc 2.0
Our group is heavily involved with the synthetic yeast genome consortium (Sc2.0, www.syntheticyeast.org), which aims to re-design and synthesize a 12 Mb designer yeast genome de novo (for details see Dymond, Nature 2010). Our group will design and synthesize at least one chromosome in house, so the success of this project will be of great significance and highly valuable to the field of synthetic biology.
Evolution of Synthetic Yeast genomes (IESY)
The Induced Evolution of Synthetic Yeast genomes (IESY) project builds on pioneering developments from the Saccharomyces cerevisiae 2.0 (Sc2.0) consortium. IESY represents an innovative concept in synthetic biology to explore combinatory diversity in pre-programmed synthetic genomes to evolve new and useful functions. Using the system for Synthetic Chromosome Rearrangement and Modification by LoxP-mediated evolution (SCRaMbLE) we are able to generate high-fitness yeast cells with synthetic chromosomes under specific conditions. The SCRaMbLE system can generate genome diversity with the capability of genome minimization and the ability to produce new biological functions through rapid evolution of genome content, copy number and gene order.
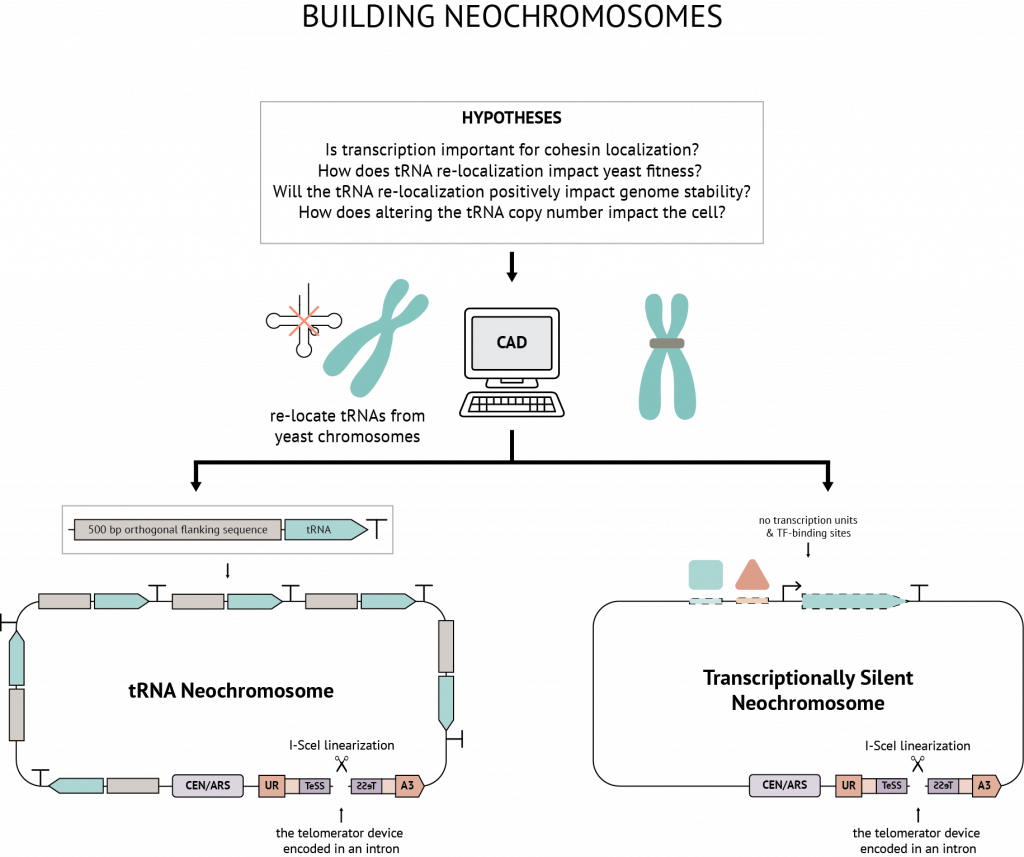
tRNA Neochromosome
This project is part of the international synthetic yeast consortium. The goal of this project is to design and build a tRNA neochromosome in yeast, hosting all of the tRNAs which will be removed from the synthetic yeast genome.
tRNA genes have been shown to be hotspots for DNA instability for several reasons. First, tRNA genes are often the target of 5’ upstream retrotransposon incorporation. Second, tRNA genes are transcribed heavily, which can lead to DNA damage caused by collisions between the replication and transcriptional machineries. Furthermore, retrotransposon insertions produce highly-homologous repeats leading to homologous recombination between chromosomal regions.
Thus, as part of Sc2.0, all tRNA genes will be moved to their own dedicated chromosome (a “Party Chromosome”). All introns will also be removed from tDNA, as another goal of the project is to observe the effect of a yeast genome with fewer introns. rox Recombination sites will be placed between the tRNA genes, which will enable us ‘SCRaMbLE’ the neochromosome and observe whether there is a preference for tRNA copy number under different conditions.